14 Advanced R
Open RStudio and then File-> New File - > R Markdown. You may be asked to install additional packages - agree to this. You will see the scripting window populated with some text - this is what R Markdown looks like.
Now press the small down arrow next to the knit button at the top of the R Markdown window and click knit to pdf. Name the file you are about to create - this will be saved in your directory. You will then see all the code chunks executed in the console - if you have any errors in your code it will stop executing and you will need to fix them and then click knit to pdf again. Once it has finished there may be a pop-up window error. If this happens select “Try Again”. If that doesn’t work you should be able to click on the file in the files list on the right.
You should have a PDF that contains the below:
14.1 R Markdown
This is an R Markdown document. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see http://rmarkdown.rstudio.com.
When you click the Knit button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this:
summary(cars)
#> speed dist
#> Min. : 4.0 Min. : 2
#> 1st Qu.:12.0 1st Qu.: 26
#> Median :15.0 Median : 36
#> Mean :15.4 Mean : 43
#> 3rd Qu.:19.0 3rd Qu.: 56
#> Max. :25.0 Max. :12014.1.1 Including Plots
You can also embed plots, for example:
These are the bulidng blocks of an R Markdown document. Note that the echo = FALSE parameter was added to the code chunk for the above plot to prevent printing of the R code that generated the plot. With echo=T the code will appear as well.
plot(pressure)
If you look in the grey area where the R code is written you will see a grey cog - click on this and you can adjust the outputs you would like to see in the final document. There is also a button that runs all code chunks above and then the green triangle runs only that code chunk. You will notice that any plots/ outputs now appear in the R Markdown window and in the console below.
On Moodle you should find the R Markdown document that created this PDF. We will use this R Markdown document to introduce functions, if/ else statements and loops in R.
First, we must set the working directory and load the practical data.
# Remember to set the working directory as usual
# Load the data. You may need to alter the file directory
Census.Data <-read.csv("worksheet_data/camden/practical_data.csv")We will also need to load the spatial data files from the previous practicals.
# load the spatial libraries
library("sp")
library("rgdal")
library("rgeos")
library("tmap")
# Load the output area shapefiles
Output.Areas <- readOGR("worksheet_data/camden", "Camden_oa11")
#> OGR data source with driver: ESRI Shapefile
#> Source: "/Users/jamestodd/Desktop/GitHub/learningR/worksheet_data/camden", layer: "Camden_oa11"
#> with 749 features
#> It has 1 fields
# join our census data to the shapefile
OA.Census <- merge(Output.Areas, Census.Data, by.x="OA11CD", by.y="OA")One of the core advantages of R and other command line packages over traditional software is the ability to automate several commands. You can tell R to repeat your methodological steps with additional variables without the need to retype the entire code again.
There are two ways of automating codes. One means is by implementing user-written functions, another is by running for loops.
14.2 User-written functions
User-written functions allow you to create a function which runs a series of commands, it can include and input and output variable. You must also name the function yourself. Once the function has been run, it is saved in your work environment by R and you can run data through it by calling it as you would do with any other function in R (I.e. myfunction(data)).
The general structure is provided below:
# to create a new function
myfunction <- function(object1, object2, ... ){
statements
return(newobject)
}
# to run the function
Output <- myfunction(data)When you create a function you do not need the name of the variables you use. You simply create a temporary object name (say ‘x’ for example), then when you run a data object through your function, it will take the place of x.
So for purposes of example, we will work on rounding our data first.
To round a variable we can simply use the round() function. It only requires us to input a data object and to specify how many decimal places we want our data rounded to.
So for one variable it would be:
This is very straight forward. We will now put this in a new function called “myfunction” which will comprise this code.
# function is called "myfunction", accepts one object (to be called x)
myfunction <- function(x){
# x is rounded to one decimal place, creates object z
z <- round(x,1)
# object z is returned
return(z)
# close function
}Now run the function, it should appear in your environment panel. To run it we simply run data through it like so:
# lets create a new data frame so we don't overwrite our original data object
newdata <- Census.Data
# runs the function, returns the output to new Qualification_2 variable in newdata
newdata$Qualificaton_2 <- myfunction(Census.Data$Qualification)This example is very simple. However, we can add further functions into our function to make it undertake more tasks. In the demonstration below, we also run logarithmic transformation prior to rounding the data.
# creates new function which logs and then rounds data
myfunction <- function(x){
z <- log(x)
y <- round(z,4)
return(y)
}
# resets the newdata object so we can run it again
newdata <- Census.Data
# runs the function
newdata$Qualification_2 <- myfunction(Census.Data$Qualification)
newdata$Unemployed_2 <- myfunction(Census.Data$Unemployed)So here we create the z variable first, then run it through a second function to produce a y variable. However, as the code is run in order, we could give both z and y the same name to overwrite them as we run the function. I.e…
myfunction <- function(x){
z <- log(x)
z <- round(z,4)
return(z)
}
Now we will try something more advanced. We will enter our mapping code using tmap into a function so we are able to create maps more easily in the future. Here we have called the function ‘map’. The code has three arguments this time (of course we could include more if we wanted to add more customisations) they are described in the table below.
| Object label | Argument |
|---|---|
| x | A polygon shapefile |
| y | A variable of the shapefile |
| z | A colour palette |
So the code is as follows…
# magic_map function with 3 arguments
magic_map <- function(x,y,z){
tm_shape(x) + tm_fill(y, palette = z, style = "quantile") + tm_borders(alpha=.4) +
tm_compass(size = 1.8, fontsize = 0.5) +
tm_layout(title = "Camden", legend.title.size = 1.1, frame = FALSE)
}Now to run the magic_map function, we can very simply call the function and enter the three input parameters (x,y,z). Running our function is an easier way to create consistent maps in the future as it requires only one small line of code to call it.
We can now use the function to create a map for any spatial polygon data frame which contains numerical data. All we have to do is call the function and enter the three arguments. There are two examples below.
# runs map function, remember we need to include all 3 arguments of the function
magic_map(OA.Census, "Unemployed", "Blues")
#> Warning: The argument fontsize of tm_compass is deprecated. It has been
#> renamed to text.size
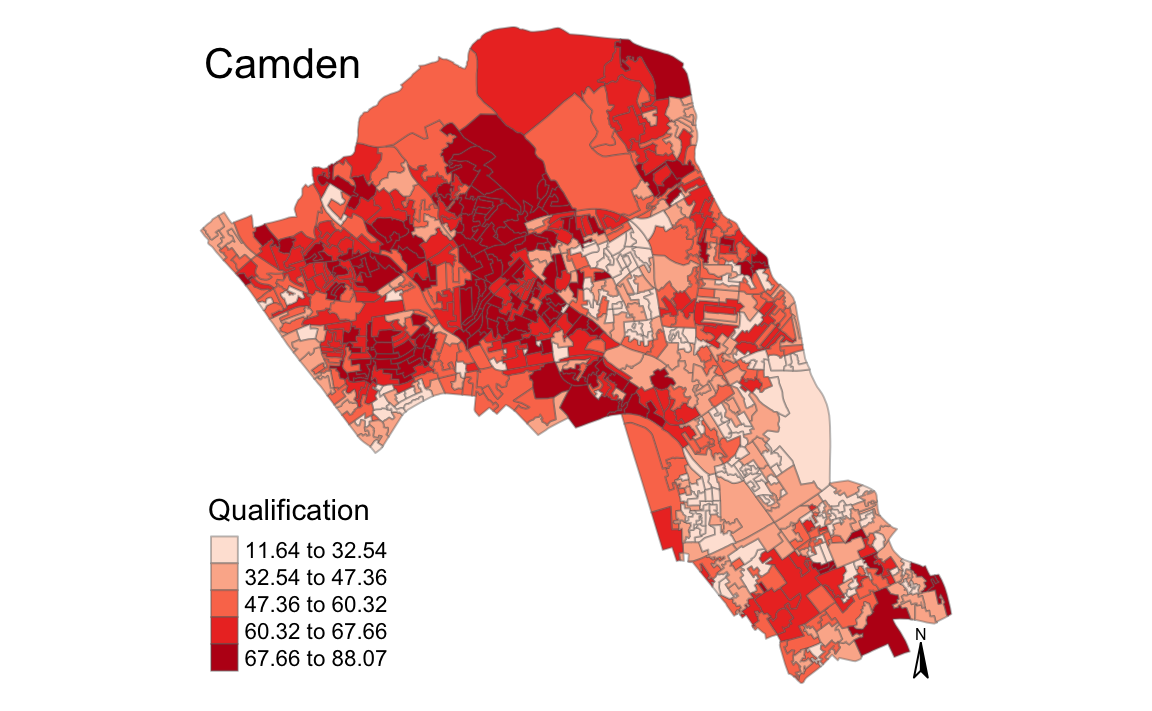
magic_map(OA.Census, "Qualification", "Reds")
#> Warning: The argument fontsize of tm_compass is deprecated. It has been
#> renamed to text.size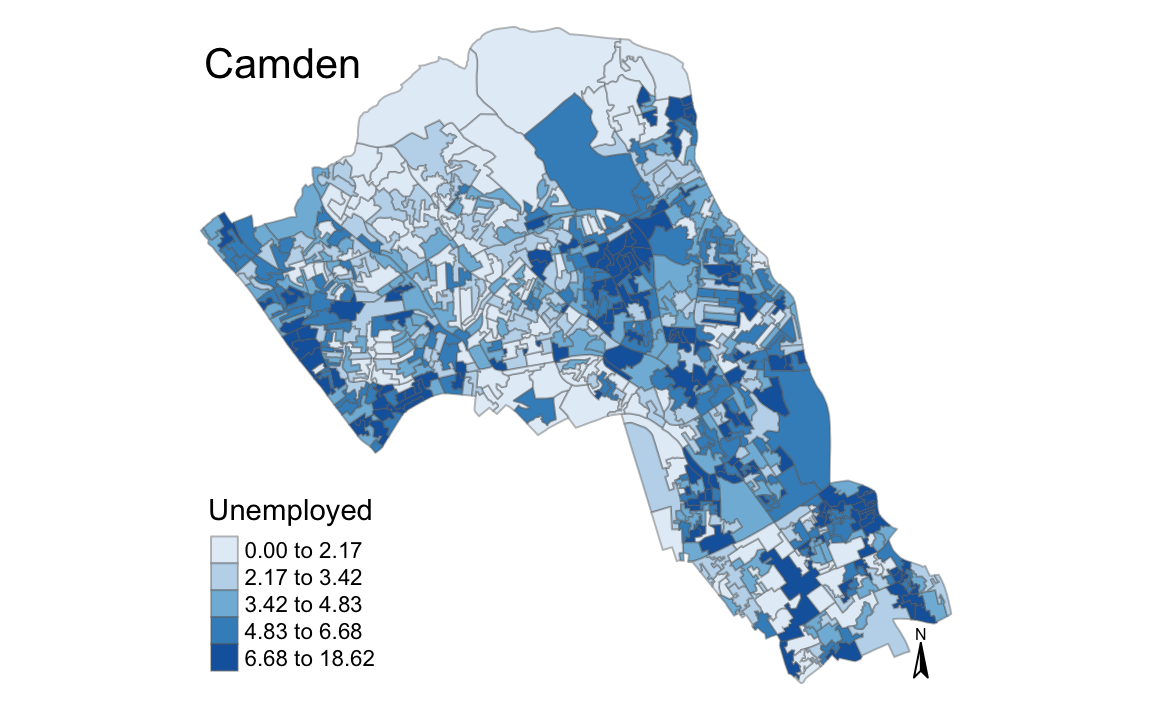
14.3 Loops
A loop is a means to repeat code under defined conditions. This is a great way for reducing the amount of code you may need to write. For example, you could use the loop to iterate a series of commands through a number of different positions (i.e. rows) in a data frame.
The most basic means of looping commands in R is known as the for loop. To implement it, you run the for() function and within the parameters, you define the sequence through which the loop will input data. Usually, an argument is assigned (i) and given a range of numerical positions (i.e. in 2:5 which runs from position 2 to position 5). You next enter the commands which the loop will run within curly brackets following the parameters. The defined argument (i) is used to determine the input data for each iteration. This has been demonstrated in the example below which loops the code for four columns of our data frame.
Note in this example we are running the loop from columns 2 onwards as column 1 does not contain our numeric data
#create a new data frame of the same properties as our data file
newdata <- Census.Data
# a for loop where i iterates from 2 to 5
for(i in 2:5){
# i is used to identify the column number
newdata[, i] <- round(Census.Data[,i], 1)
}We can also use the ncol() function if you do not know the total number of columns or accept that the number may change under certain circumstances.
# use ncol to determine the number of columns
for(i in 2: ncol (Census.Data)){
newdata[, i] <- Census.Data[,i]/100
}We will be using the ratio data from newdata in the upcoming section.
14.4 If. else statements
There are various commands we can run within for loops to edit their functionality. If-else statements allow us to change the outcome of the loop depending on a particular statement. For instance, if a value is below a certain threshold, you can tell the loop to round it up, otherwise (else) do nothing.
This time we are going to run the loop for every row in one variable to recode them to “low” and “high” labels. In this example, we are asking if the value in our data is below 0.5. If it is then we are giving it a “low” label. The else argument determines what R will do if the value does not meet the criterion, in this case it is given a “high” label.
# creates a new newdata object so we have it saved somewhere
newdata1 <- newdata
for(i in 1:nrow(newdata)){
if (newdata$White_British[i] < 0.5) {
newdata$White_British[i] <- "Low";
} else {
newdata$White_British[i] <- "High";
}
}We can also input multiple else statements.
# copies the numbers back to newdata so we can start again
newdata <- newdata1
for(i in 1:nrow(newdata)){
if (newdata$White_British[i] < 0.25) {
newdata$White_British[i] <- "Very Low";
} else if (newdata$White_British[i] < 0.50){
newdata$White_British[i] <- "Low";
} else if (newdata$White_British[i] < 0.75){
newdata$White_British[i] <- "High";
} else {
newdata$White_British[i] <- "Very High";
}
}We can also nest multiple loops if we wish to loop through two different arguments. In this example, we are going to loop both the columns and rows for our newdata data frame. This will reassign all of our variables in the data into four interval labels.
# copies the numbers back to newdata so we can start again
newdata <- newdata1
for(j in 2: ncol (newdata)){
for(i in 1:nrow(newdata)){
if (newdata[i,j] < 0.25) {
newdata[i,j] <- "Very Low";
} else if (newdata[i,j] < 0.50){
newdata[i,j] <- "Low";
} else if (newdata[i,j] < 0.75){
newdata[i,j] <- "High";
} else {
newdata[i,j] <- "Very High";
}
}
}We can join the newdata data frame to our output area shapefile to map it using our earlier generated magic_map function.
# merge our new formatted data with the output areas shapefile
shapefile <- merge(Output.Areas, newdata, by.x = "OA11CD", by.y = "OA")
# runs our predefined map function
magic_map(shapefile, "Qualification", "Set2")
#> Warning: The argument fontsize of tm_compass is deprecated. It has been
#> renamed to text.size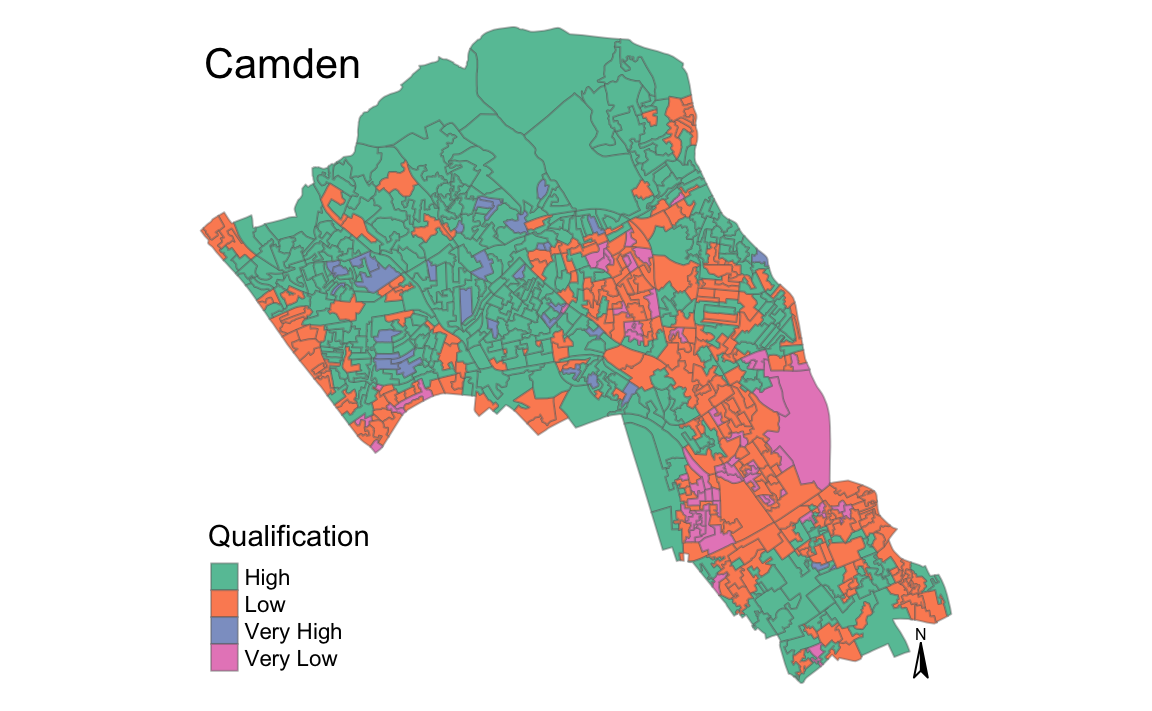
Now press the small down arrow next to the knit button at the top of the R Markdown window and click knit to pdf. Name the file you are about to create - this will be saved in your directory. You will then see all the code chunks executed in the console - if you have any errors in your code it will stop executing and you will need to fix them and then click knit to pdf again. Once it has finished there may be a pop-up window error. If this happens select “Try Again”. If that doesn’t work you should be able to click on the file in the files list on the right.
14.5 Creating Dot Density Map with a Loop
Here I detail how to create a dot density map of the population in Britain by looping through the plot command. Please note this approach can create a large file so please follow the steps carefully if you want to avoid system crashes!
First of all we need to load in the population grid for Great Britain. Each grid cell has a population value associated with it, and it is that
GB_Pop_Grid can be downloaded from here https://dataviz.spatial.ly/worksheets/gb_pop_grid.zip and should be placed in your working directory.
library(rgdal)
#GB_Pop_Grid can be downloaded from here
Input<-readOGR(dsn="worksheet_data/gb_pop_grid", layer="GB_Pop_Grid")
#> OGR data source with driver: ESRI Shapefile
#> Source: "/Users/jamestodd/Desktop/GitHub/learningR/worksheet_data/gb_pop_grid", layer: "GB_Pop_Grid"
#> with 119622 features
#> It has 3 fieldsNext we create an empty PNG file in the working directly- this is my preferred image format but you could also use JPEG or TIFF. A PDF is a bad idea here since theres going to be a lot of points plotted and it will struggle to handle them.
#Open PNG- the dimensions are 50cm by 100cm with a resolution of 150 - so it's a big image!
png("GB_density.png",width=50, height=100, units="cm", res=150)Next we use the par function to set our plot background to black.
par(bg='black')We can then use the bbox function to get the four corners that form the spatial extent of the grid data. This can be wrapped in the plot function to create the blank plot upon which we will overlay the points. Use the help files to check what the asp, axes and ann parameters are for.
plot(t(bbox(Input)), asp=T, axes=F, ann=F)Now we’re ready for the loop!
There are 4 steps: 1. for (i in 1:nrow(Input)) This is the start of the loop and essentially tells R to assign a new value to i for each iteration of our loop. In this case i is assigned every value between 1 and the number of of rows that the input object has. For example if input has 10 rows i will first be 1 and then 2 and then 3 and so on until 10. 2. Poly<- Input[i,] This creates a new object Poly from a subset of the Input object. This is the crucial step because i is changing with each iteration. So in practice this means we are effectively running Poly<- Input[1,], Poly<- Input[2,],Poly<- Input[3,] and so on until Poly<- Input[10,] (if Input has 10 rows) 3. pts1<-spsample(Poly, n = 1+round(Poly@data$Pop/1000), "random", iter=15) This step uses the spsample function to generate our points that we wish to plot. Use the help files to establish what the parameters are. As a clue - the above code will create 1 point for every 1000 people within each grid cell. If you reduce this to say 1 dot per 100 people it will use a lot of computing resource so best keep this number above 500 if using the RStudio server. 4. plot(pts1, pch=16, cex=0.03, add=T, col="cyan") Here we plot the points generated. 5. print(i) Print i to show the progress of the loop.
#Loop through each grid square and fill with dots.
for (i in 1:nrow(Input))
{
Poly<- Input[i,]
pts1<-spsample(Poly, n = 1+round(Poly@data$Pop/1000), "random", iter=15)
plot(pts1, pch=16, cex=0.03, add=T, col="cyan")
print(i)
}
dev.off()